Exploration
Loading required packages and cleaned data.
#load required packages
library(tidyverse)
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.0 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.1 ✔ tibble 3.1.8
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.1
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
here() starts at C:/Users/Katie/Documents/2022-2023/MADA/katiewells-MADA-portfolio
#load data
flu2 <- readRDS(here("fluanalysis", "data", "flu2.rds"))
Summary statistics for BodyTemp and Nausea
#provide summary data for important variables
flu2 %>% pull(Nausea) %>% summary()
flu2 %>% pull(BodyTemp) %>% summary()
Min. 1st Qu. Median Mean 3rd Qu. Max.
97.20 98.20 98.50 98.94 99.30 103.10
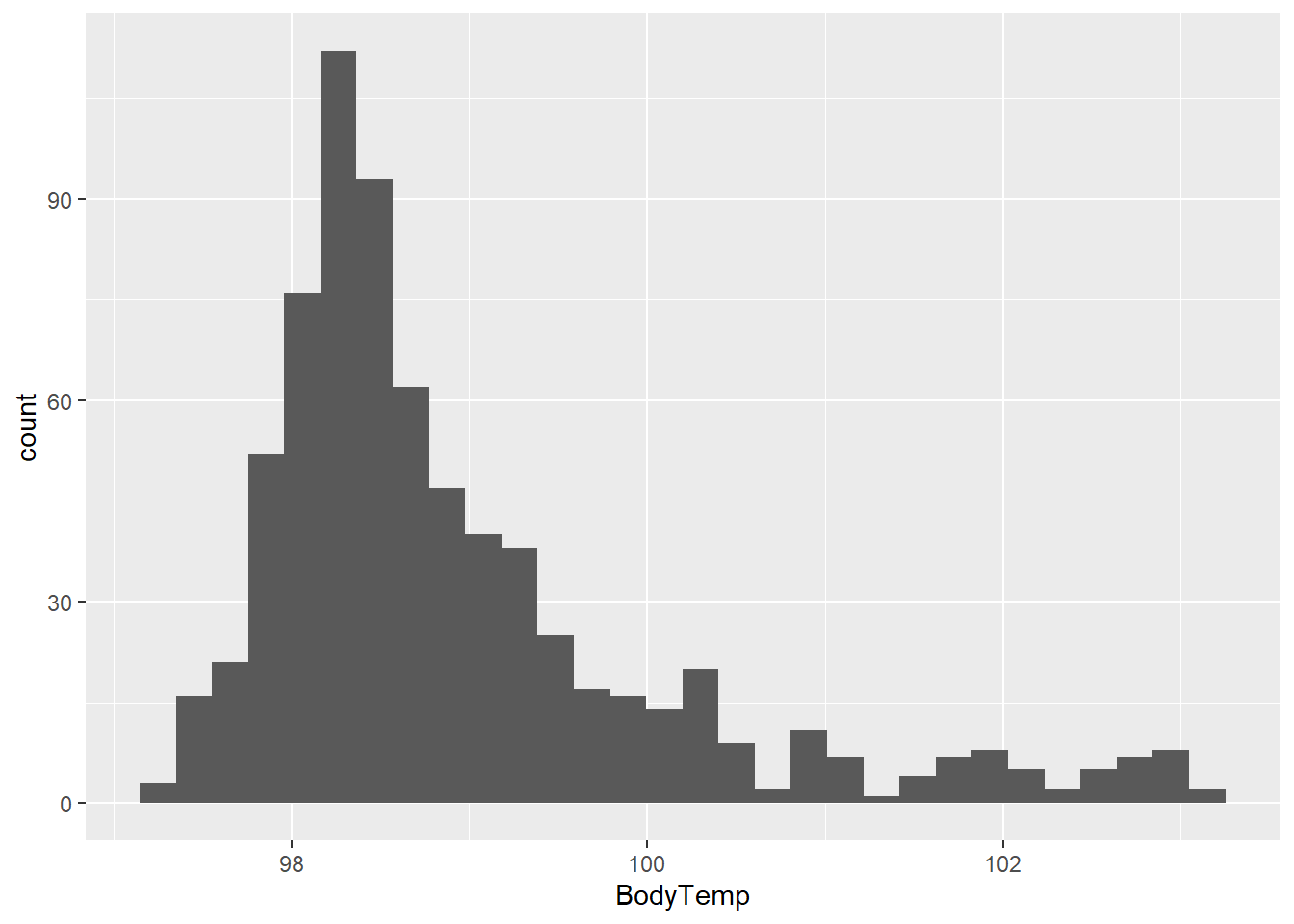
Distribution of BodyTemp
#look at the distribution of BodyTemp
flu2 %>% ggplot(aes(x=BodyTemp)) + geom_histogram()
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Looks like of the body temperatures cluster around 98.5 degrees with some right skew.
Lets take a look ath the relationship between BodyTemp and some predictors.
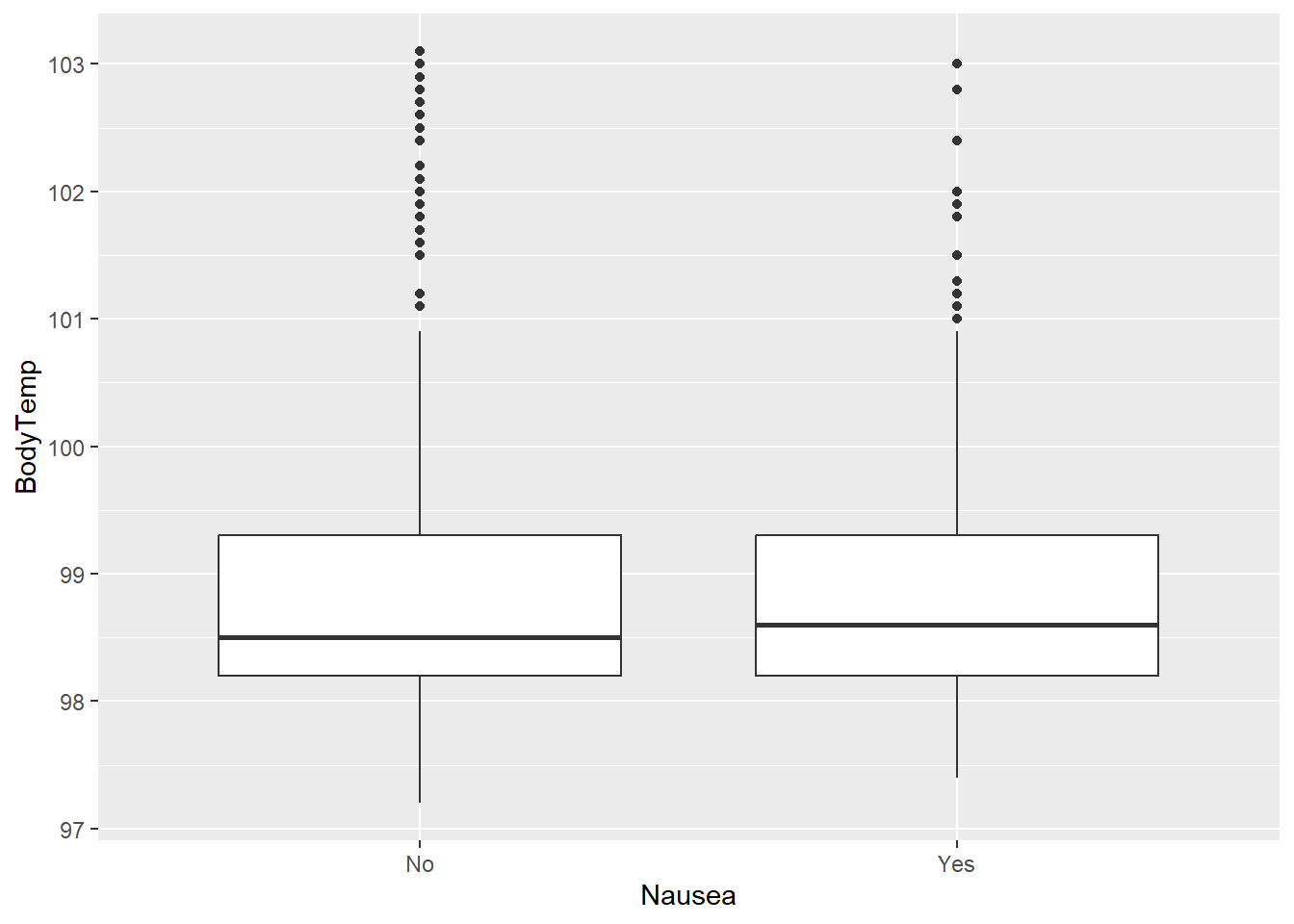
flu2 %>% ggplot(aes(x=Nausea, y=BodyTemp)) + geom_boxplot()
Seems like median body temperature is just slightly higher in those with nausea than without.
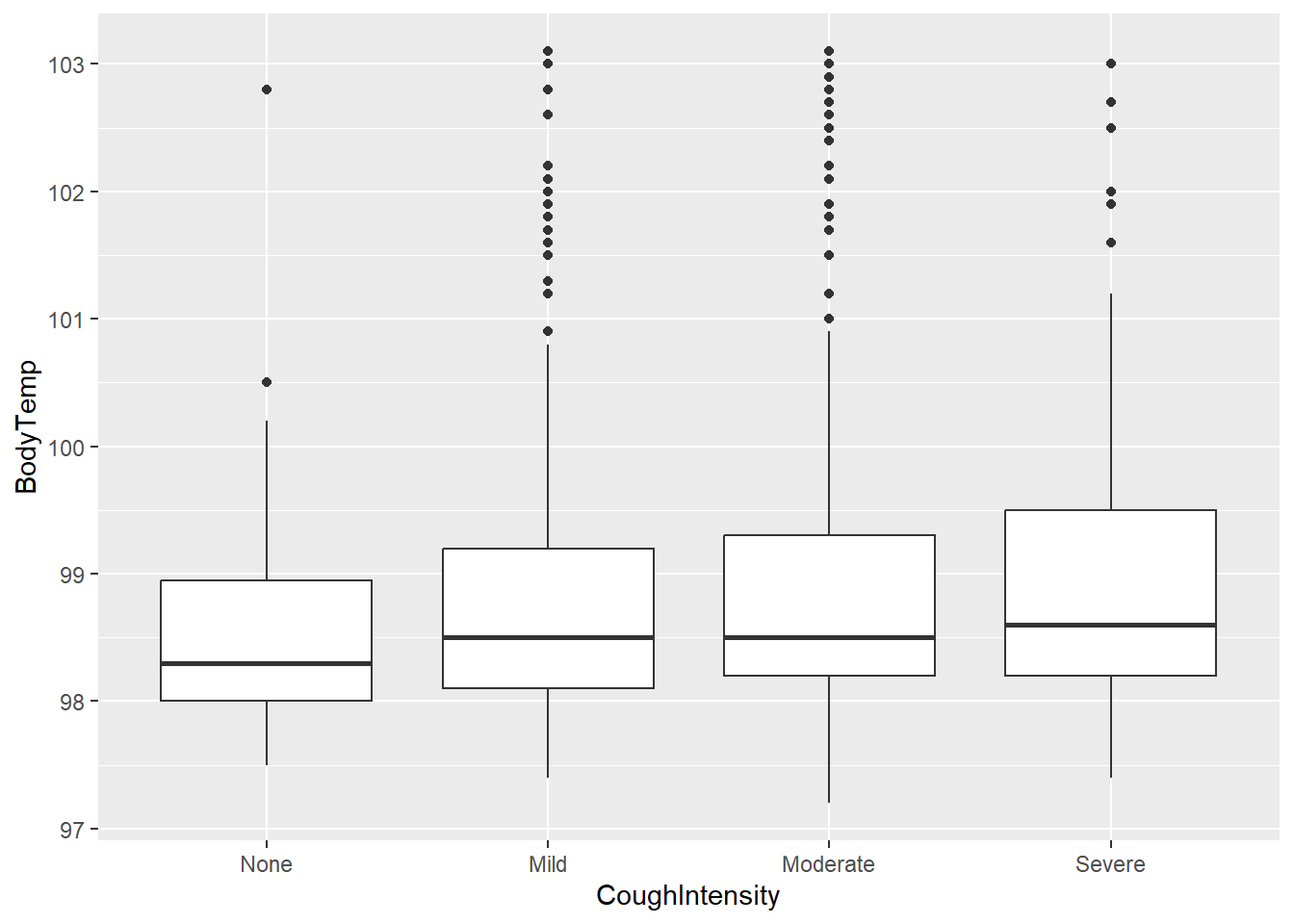
flu2 %>% ggplot(aes(x=CoughIntensity, y=BodyTemp)) + geom_boxplot()
Median body temperature looks to increase as cough intensity increases.
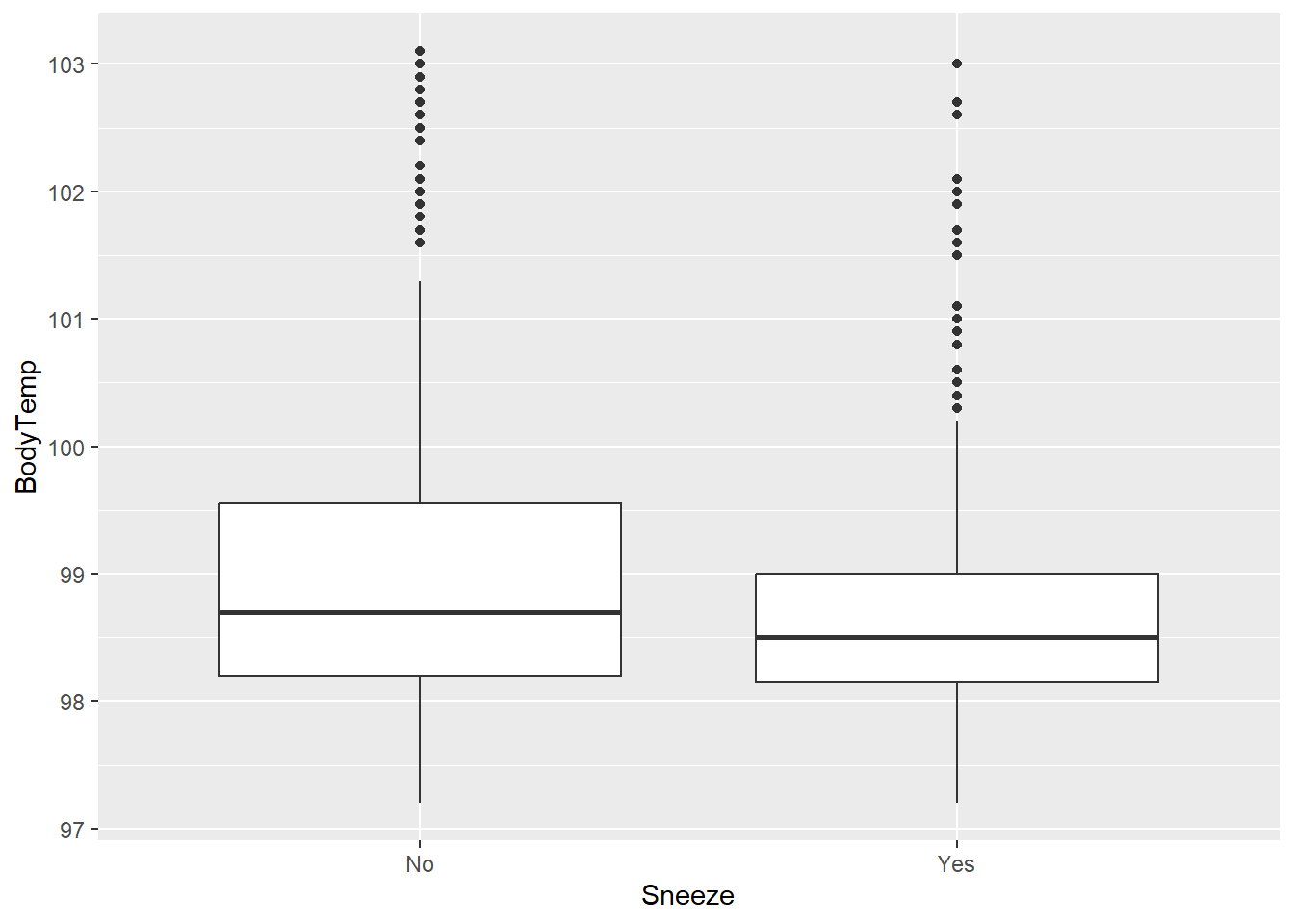
flu2 %>% ggplot(aes(x=Sneeze, y=BodyTemp)) + geom_boxplot()
Looks like people who did not report sneezing have a higher median body temperature. Weird.
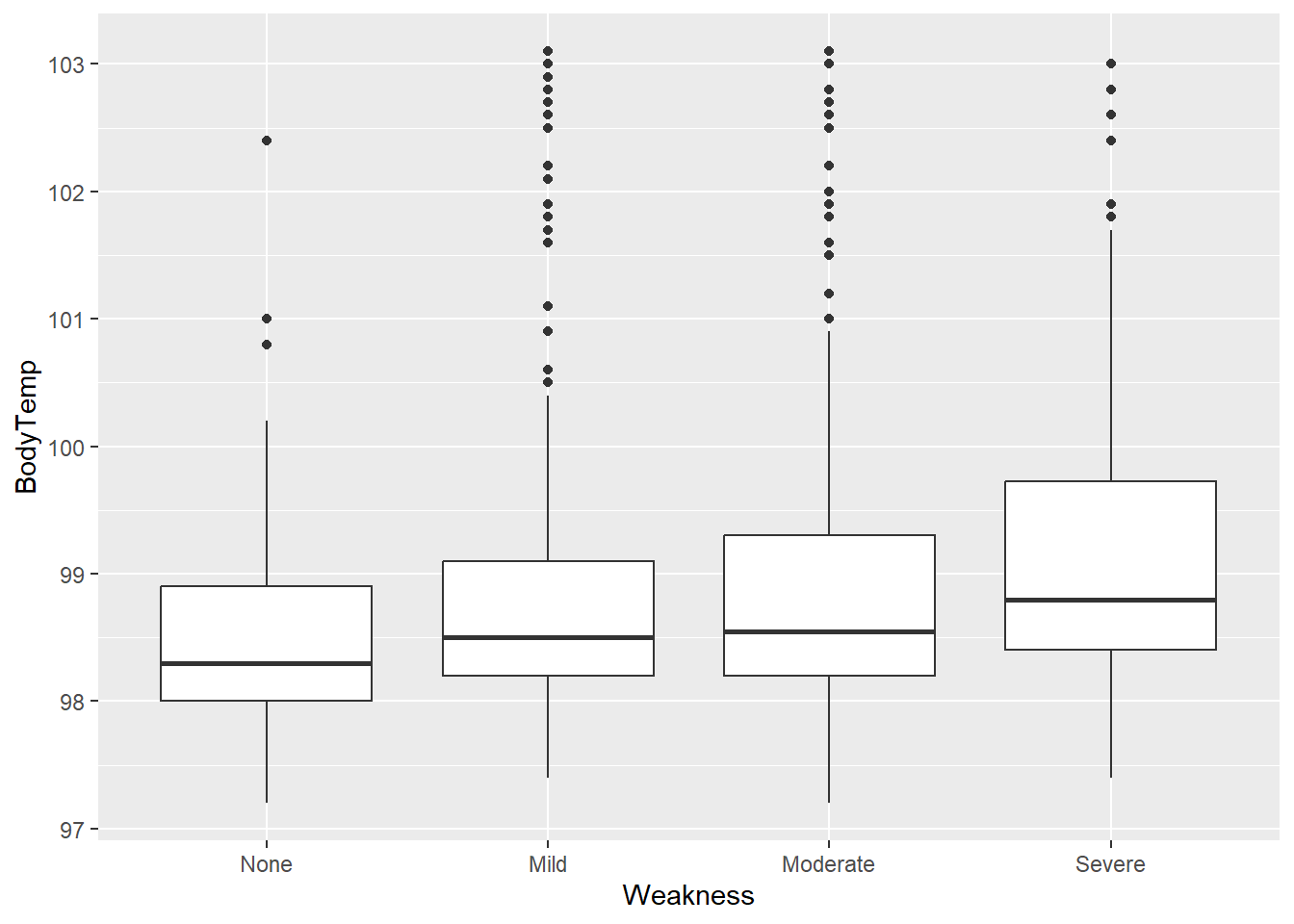
flu2 %>% ggplot(aes(x=Weakness, y=BodyTemp)) + geom_boxplot()
Median body temperature seems to increase as weakness increases.