#load required packages
library(tidyverse)
library(here)
library(tidymodels)
library(ranger)
library(glmnet)
library(rpart.plot)
library(vip)Machine Learning
Loading the needed packages and the raw data.
# load cleaned data
flu2 <- readRDS(here("fluanalysis", "data", "flu2.rds"))Setup
#set seed
set.seed(123)
#split data
split <- initial_split(flu2, strata = BodyTemp, prop = 7/10)
#create test and train datasets
flu_train1 <- training(split)
flu_test1 <- testing(split)#cross-validation 5 x 5
folds_train <- vfold_cv(flu_train1, v = 5, repeats = 5, strata = BodyTemp)
folds_train# 5-fold cross-validation repeated 5 times using stratification
# A tibble: 25 × 3
splits id id2
<list> <chr> <chr>
1 <split [405/103]> Repeat1 Fold1
2 <split [405/103]> Repeat1 Fold2
3 <split [406/102]> Repeat1 Fold3
4 <split [408/100]> Repeat1 Fold4
5 <split [408/100]> Repeat1 Fold5
6 <split [405/103]> Repeat2 Fold1
7 <split [405/103]> Repeat2 Fold2
8 <split [406/102]> Repeat2 Fold3
9 <split [408/100]> Repeat2 Fold4
10 <split [408/100]> Repeat2 Fold5
# … with 15 more rowsfolds_test <- vfold_cv(flu_test1, v = 5, repeats = 5, strata = BodyTemp)
folds_test# 5-fold cross-validation repeated 5 times using stratification
# A tibble: 25 × 3
splits id id2
<list> <chr> <chr>
1 <split [175/47]> Repeat1 Fold1
2 <split [176/46]> Repeat1 Fold2
3 <split [179/43]> Repeat1 Fold3
4 <split [179/43]> Repeat1 Fold4
5 <split [179/43]> Repeat1 Fold5
6 <split [175/47]> Repeat2 Fold1
7 <split [176/46]> Repeat2 Fold2
8 <split [179/43]> Repeat2 Fold3
9 <split [179/43]> Repeat2 Fold4
10 <split [179/43]> Repeat2 Fold5
# … with 15 more rowsMaking recipe
#recipe creation
flu_rec <- recipe(BodyTemp ~ ., data = flu_train1) %>%
step_dummy(all_nominal(), -all_outcomes()) Null model
#null model
null_mod <- null_model() %>%
set_engine("parsnip") %>%
set_mode("regression")training data
#null model recipe with training data
null_recipe_train <- recipe(BodyTemp ~ 1, data = flu_train1)
null_wf_train <- workflow() %>% add_model(null_mod) %>% add_recipe(null_recipe_train)
null_train_fit <-
fit_resamples(null_wf_train, resamples = folds_train)testing data
#null model recipe with testing data
null_recipe_test <- recipe(BodyTemp ~ 1, data = flu_test1)
null_wf_test <- workflow() %>% add_model(null_mod) %>% add_recipe(null_recipe_test)
null_test_fit <-
fit_resamples(null_wf_test, resamples = folds_test)#collect metrics from null
null_train_fit %>% collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 1.21 25 0.0177 Preprocessor1_Model1
2 rsq standard NaN 0 NA Preprocessor1_Model1null_test_fit %>% collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 1.16 25 0.0285 Preprocessor1_Model1
2 rsq standard NaN 0 NA Preprocessor1_Model1Model Tuning and Fitting
Tree
Model Specification
tune_spec <- decision_tree(cost_complexity = tune(),
tree_depth = tune()) %>%
set_engine("rpart") %>%
set_mode("regression")
tune_specDecision Tree Model Specification (regression)
Main Arguments:
cost_complexity = tune()
tree_depth = tune()
Computational engine: rpart Workflow Definition
tree_wf <- workflow() %>%
add_model(tune_spec) %>%
add_recipe(flu_rec)Tuning Grid Specification
tree_grid <- grid_regular(cost_complexity(),
tree_depth(),
levels = 5)
tree_grid# A tibble: 25 × 2
cost_complexity tree_depth
<dbl> <int>
1 0.0000000001 1
2 0.0000000178 1
3 0.00000316 1
4 0.000562 1
5 0.1 1
6 0.0000000001 4
7 0.0000000178 4
8 0.00000316 4
9 0.000562 4
10 0.1 4
# … with 15 more rowstree_grid %>%
count(tree_depth)# A tibble: 5 × 2
tree_depth n
<int> <int>
1 1 5
2 4 5
3 8 5
4 11 5
5 15 5Tuning using Cross-Validation and Tune_grid()
tree_res <- tree_wf %>%
tune_grid(
resamples = folds_train,
grid = tree_grid
)! Fold1, Repeat1: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold2, Repeat1: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold3, Repeat1: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold4, Repeat1: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold5, Repeat1: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold1, Repeat2: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold2, Repeat2: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold3, Repeat2: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold4, Repeat2: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold5, Repeat2: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold1, Repeat3: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold2, Repeat3: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold3, Repeat3: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold4, Repeat3: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold5, Repeat3: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold1, Repeat4: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold2, Repeat4: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold3, Repeat4: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold4, Repeat4: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold5, Repeat4: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold1, Repeat5: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold2, Repeat5: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold3, Repeat5: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold4, Repeat5: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...! Fold5, Repeat5: internal:
There was 1 warning in `dplyr::summarise()`.
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 1`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 4`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 8`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 11`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...
ℹ In argument: `.estimate = metric_fn(truth = BodyTemp, estimate = .pr...
= na_rm)`.
ℹ In group 1: `cost_complexity = 0.1`, `tree_depth = 15`.
Caused by warning:
! A correlation computation is required, but `estimate` is constant an...tree_res# Tuning results
# 5-fold cross-validation repeated 5 times using stratification
# A tibble: 25 × 5
splits id id2 .metrics .notes
<list> <chr> <chr> <list> <list>
1 <split [405/103]> Repeat1 Fold1 <tibble [50 × 6]> <tibble [1 × 3]>
2 <split [405/103]> Repeat1 Fold2 <tibble [50 × 6]> <tibble [1 × 3]>
3 <split [406/102]> Repeat1 Fold3 <tibble [50 × 6]> <tibble [1 × 3]>
4 <split [408/100]> Repeat1 Fold4 <tibble [50 × 6]> <tibble [1 × 3]>
5 <split [408/100]> Repeat1 Fold5 <tibble [50 × 6]> <tibble [1 × 3]>
6 <split [405/103]> Repeat2 Fold1 <tibble [50 × 6]> <tibble [1 × 3]>
7 <split [405/103]> Repeat2 Fold2 <tibble [50 × 6]> <tibble [1 × 3]>
8 <split [406/102]> Repeat2 Fold3 <tibble [50 × 6]> <tibble [1 × 3]>
9 <split [408/100]> Repeat2 Fold4 <tibble [50 × 6]> <tibble [1 × 3]>
10 <split [408/100]> Repeat2 Fold5 <tibble [50 × 6]> <tibble [1 × 3]>
# … with 15 more rows
There were issues with some computations:
- Warning(s) x25: There was 1 warning in `dplyr::summarise()`. ℹ In argument: `.est...
Run `show_notes(.Last.tune.result)` for more information.tree_res %>%
collect_metrics()# A tibble: 50 × 8
cost_complexity tree_depth .metric .estimator mean n std_err .config
<dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0000000001 1 rmse standard 1.19 25 0.0181 Prepro…
2 0.0000000001 1 rsq standard 0.0361 25 0.00422 Prepro…
3 0.0000000178 1 rmse standard 1.19 25 0.0181 Prepro…
4 0.0000000178 1 rsq standard 0.0361 25 0.00422 Prepro…
5 0.00000316 1 rmse standard 1.19 25 0.0181 Prepro…
6 0.00000316 1 rsq standard 0.0361 25 0.00422 Prepro…
7 0.000562 1 rmse standard 1.19 25 0.0181 Prepro…
8 0.000562 1 rsq standard 0.0361 25 0.00422 Prepro…
9 0.1 1 rmse standard 1.21 25 0.0177 Prepro…
10 0.1 1 rsq standard NaN 0 NA Prepro…
# … with 40 more rows#plot using autoplot
tree_res %>% autoplot()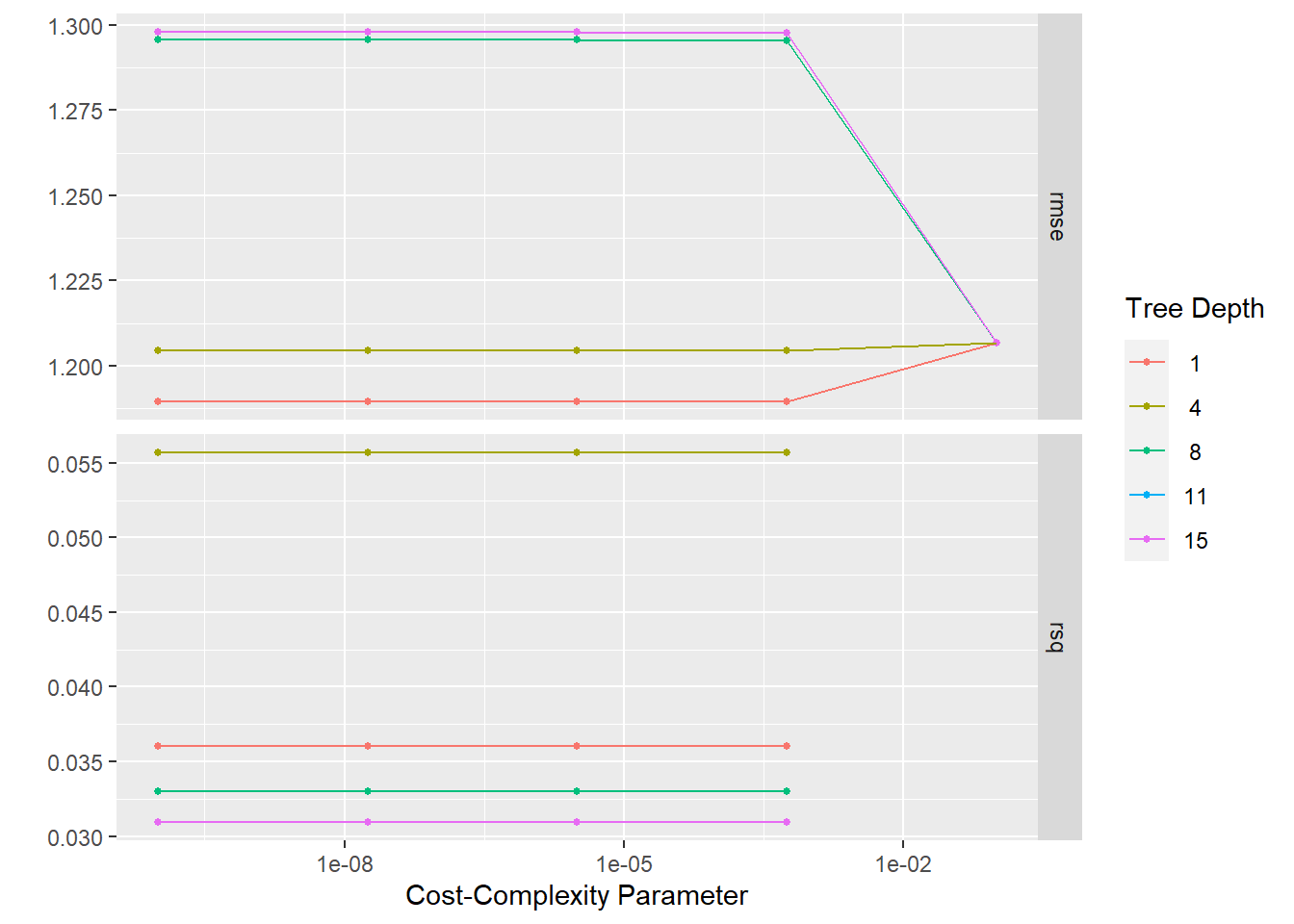
#getting best model
tree_res %>%
show_best(metric = "rmse")# A tibble: 5 × 8
cost_complexity tree_depth .metric .estimator mean n std_err .config
<dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0000000001 1 rmse standard 1.19 25 0.0181 Preprocesso…
2 0.0000000178 1 rmse standard 1.19 25 0.0181 Preprocesso…
3 0.00000316 1 rmse standard 1.19 25 0.0181 Preprocesso…
4 0.000562 1 rmse standard 1.19 25 0.0181 Preprocesso…
5 0.0000000001 4 rmse standard 1.20 25 0.0182 Preprocesso…best_tree <- tree_res %>%
select_best(metric = "rmse")
best_tree# A tibble: 1 × 3
cost_complexity tree_depth .config
<dbl> <int> <chr>
1 0.0000000001 1 Preprocessor1_Model01Finalizing fit with best model
#finalizing workflows with best models
final_tree_wf <- tree_wf %>%
finalize_workflow(best_tree)
final_tree_fit <- final_tree_wf %>% fit(data=flu_train1)
final_tree_fit══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: decision_tree()
── Preprocessor ────────────────────────────────────────────────────────────────
1 Recipe Step
• step_dummy()
── Model ───────────────────────────────────────────────────────────────────────
n= 508
node), split, n, deviance, yval
* denotes terminal node
1) root 508 742.9363 98.93642
2) Sneeze_Yes>=0.5 280 259.6477 98.69107 *
3) Sneeze_Yes< 0.5 228 445.7356 99.23772 *#plot tree
rpart.plot(extract_fit_parsnip(final_tree_fit)$fit)Warning: Cannot retrieve the data used to build the model (so cannot determine roundint and is.binary for the variables).
To silence this warning:
Call rpart.plot with roundint=FALSE,
or rebuild the rpart model with model=TRUE.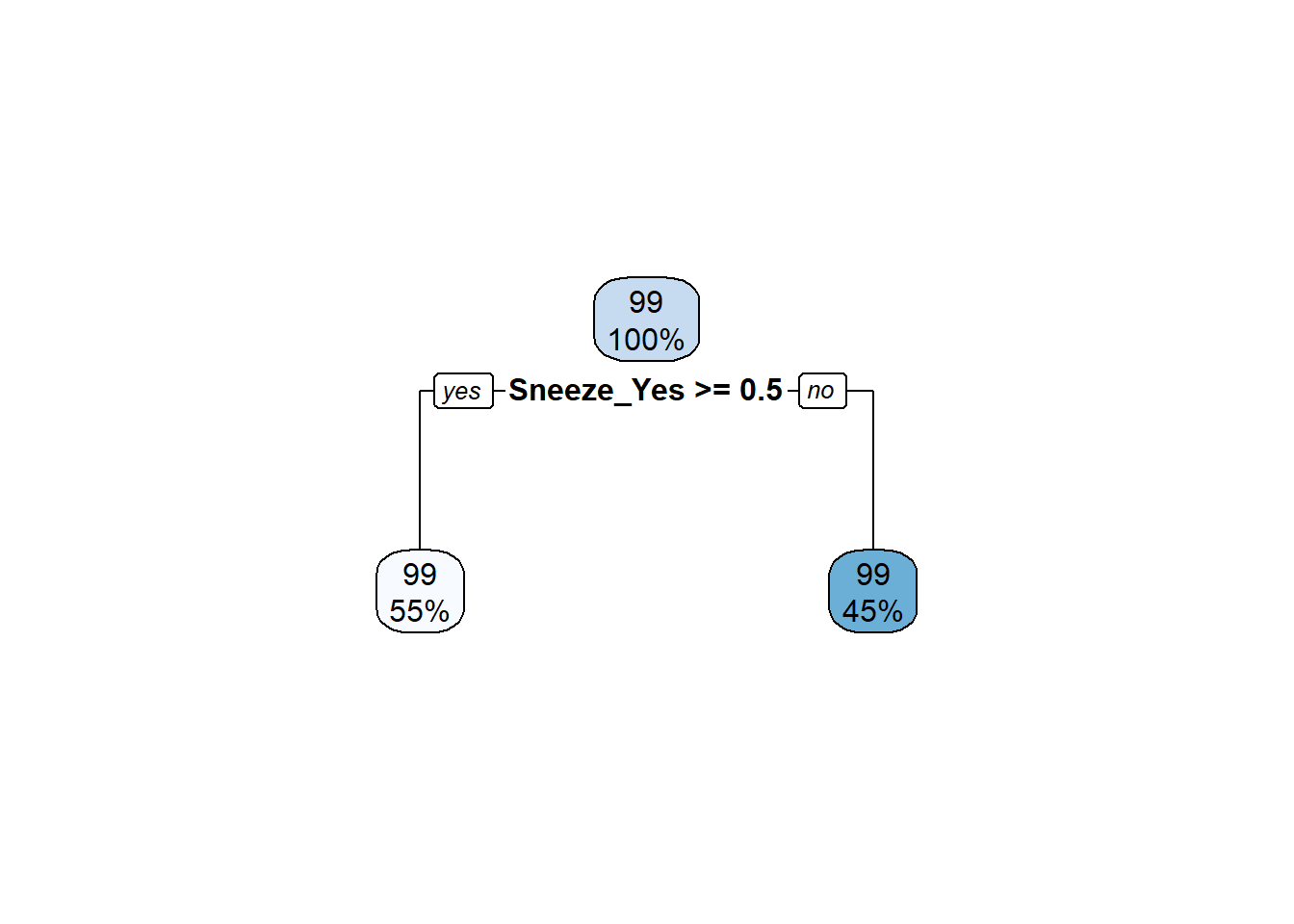
Lasso
Model Specification
lr_mod <- linear_reg(penalty = tune(), mixture = 1) %>%
set_engine("glmnet")Workflow Definition
lr_workflow <- workflow() %>%
add_model(lr_mod) %>%
add_recipe(flu_rec)Tuning Grid Specification
lr_reg_grid <- tibble(penalty = 10^seq(-4, -1, length.out = 30))
lr_reg_grid %>% top_n(-5)Selecting by penalty# A tibble: 5 × 1
penalty
<dbl>
1 0.0001
2 0.000127
3 0.000161
4 0.000204
5 0.000259lr_reg_grid %>% top_n(5)Selecting by penalty# A tibble: 5 × 1
penalty
<dbl>
1 0.0386
2 0.0489
3 0.0621
4 0.0788
5 0.1 Tuning using cross-validation and the tune_grid() function
lr_res <- lr_workflow %>%
tune_grid(resamples = folds_train,
grid = lr_reg_grid,
control = control_grid(verbose = FALSE, save_pred = TRUE),
metrics = metric_set(rmse))
lr_res %>% collect_metrics()# A tibble: 30 × 7
penalty .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0001 rmse standard 1.18 25 0.0167 Preprocessor1_Model01
2 0.000127 rmse standard 1.18 25 0.0167 Preprocessor1_Model02
3 0.000161 rmse standard 1.18 25 0.0167 Preprocessor1_Model03
4 0.000204 rmse standard 1.18 25 0.0167 Preprocessor1_Model04
5 0.000259 rmse standard 1.18 25 0.0167 Preprocessor1_Model05
6 0.000329 rmse standard 1.18 25 0.0167 Preprocessor1_Model06
7 0.000418 rmse standard 1.18 25 0.0167 Preprocessor1_Model07
8 0.000530 rmse standard 1.18 25 0.0167 Preprocessor1_Model08
9 0.000672 rmse standard 1.18 25 0.0167 Preprocessor1_Model09
10 0.000853 rmse standard 1.18 25 0.0167 Preprocessor1_Model10
# … with 20 more rows#plot using autoplot
lr_res %>% autoplot()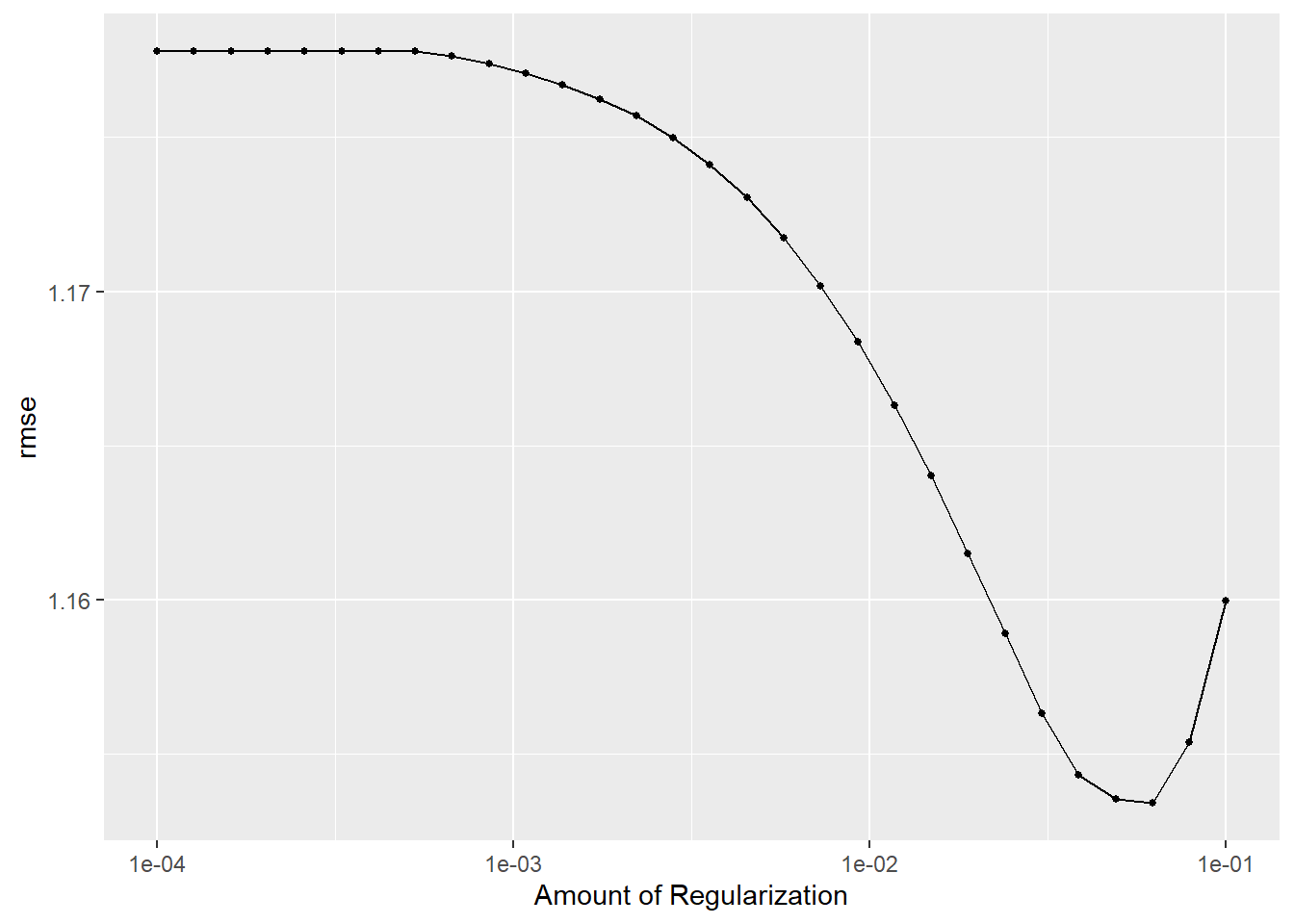
#getting best model
lr_res %>%
show_best(metric = "rmse")# A tibble: 5 × 7
penalty .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0621 rmse standard 1.15 25 0.0169 Preprocessor1_Model28
2 0.0489 rmse standard 1.15 25 0.0169 Preprocessor1_Model27
3 0.0386 rmse standard 1.15 25 0.0169 Preprocessor1_Model26
4 0.0788 rmse standard 1.16 25 0.0171 Preprocessor1_Model29
5 0.0304 rmse standard 1.16 25 0.0169 Preprocessor1_Model25best_lr <- lr_res %>%
select_best(metric = "rmse")
best_lr# A tibble: 1 × 2
penalty .config
<dbl> <chr>
1 0.0621 Preprocessor1_Model28#finalizing workflows with best models
final_lr_wf <- lr_workflow %>%
finalize_workflow(best_lr)
final_lr_fit <- final_lr_wf %>% fit(data=flu_train1)
final_lr_fit══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: linear_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
1 Recipe Step
• step_dummy()
── Model ───────────────────────────────────────────────────────────────────────
Call: glmnet::glmnet(x = maybe_matrix(x), y = y, family = "gaussian", alpha = ~1)
Df %Dev Lambda
1 0 0.00 0.271900
2 2 1.24 0.247700
3 2 2.67 0.225700
4 2 3.86 0.205700
5 2 4.85 0.187400
6 2 5.67 0.170800
7 2 6.35 0.155600
8 2 6.91 0.141800
9 5 7.57 0.129200
10 5 8.27 0.117700
11 8 9.06 0.107200
12 8 9.81 0.097710
13 9 10.44 0.089030
14 9 11.09 0.081120
15 9 11.63 0.073920
16 10 12.12 0.067350
17 10 12.56 0.061370
18 12 13.00 0.055910
19 14 13.45 0.050950
20 16 13.85 0.046420
21 19 14.24 0.042300
22 19 14.59 0.038540
23 19 14.87 0.035120
24 22 15.17 0.032000
25 22 15.44 0.029150
26 22 15.67 0.026560
27 22 15.85 0.024200
28 23 16.01 0.022050
29 24 16.15 0.020090
30 25 16.28 0.018310
31 25 16.39 0.016680
32 25 16.49 0.015200
33 25 16.56 0.013850
34 26 16.63 0.012620
35 27 16.69 0.011500
36 27 16.73 0.010480
37 27 16.77 0.009547
38 27 16.81 0.008698
39 28 16.84 0.007926
40 29 16.86 0.007222
41 29 16.88 0.006580
42 29 16.90 0.005995
43 29 16.91 0.005463
44 29 16.92 0.004978
45 30 16.93 0.004535
46 30 16.94 0.004132
...
and 22 more lines.x <- final_lr_fit$fit$fit$fit
plot(x, "lambda")
Random Forest
Model Specification
cores <- parallel::detectCores()
cores[1] 8rf_mod <-
rand_forest(mtry = tune(), min_n = tune(), trees = 1000) %>%
set_engine("ranger", num.threads = cores) %>%
set_mode("regression")Workflow Definition
rf_workflow <-
workflow() %>%
add_model(rf_mod) %>%
add_recipe(flu_rec)Tuning Grid Specification
rf_modRandom Forest Model Specification (regression)
Main Arguments:
mtry = tune()
trees = 1000
min_n = tune()
Engine-Specific Arguments:
num.threads = cores
Computational engine: ranger extract_parameter_set_dials(rf_mod)Collection of 2 parameters for tuning
identifier type object
mtry mtry nparam[?]
min_n min_n nparam[+]
Model parameters needing finalization:
# Randomly Selected Predictors ('mtry')
See `?dials::finalize` or `?dials::update.parameters` for more information.Tuning Using Cross-Validation and the Tune_Grid()
rf_res <- rf_workflow %>%
tune_grid(resamples = folds_train,
grid = 25,
control = control_grid(save_pred = TRUE),
metrics = metric_set(rmse))i Creating pre-processing data to finalize unknown parameter: mtryrf_res %>%
collect_metrics()# A tibble: 25 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 13 13 rmse standard 1.19 25 0.0165 Preprocessor1_Model01
2 5 36 rmse standard 1.16 25 0.0166 Preprocessor1_Model02
3 16 28 rmse standard 1.18 25 0.0164 Preprocessor1_Model03
4 30 40 rmse standard 1.18 25 0.0167 Preprocessor1_Model04
5 11 30 rmse standard 1.17 25 0.0163 Preprocessor1_Model05
6 7 26 rmse standard 1.17 25 0.0166 Preprocessor1_Model06
7 22 26 rmse standard 1.19 25 0.0167 Preprocessor1_Model07
8 10 11 rmse standard 1.19 25 0.0163 Preprocessor1_Model08
9 7 2 rmse standard 1.19 25 0.0164 Preprocessor1_Model09
10 9 6 rmse standard 1.19 25 0.0162 Preprocessor1_Model10
# … with 15 more rows#plot with autoplot
autoplot(rf_res)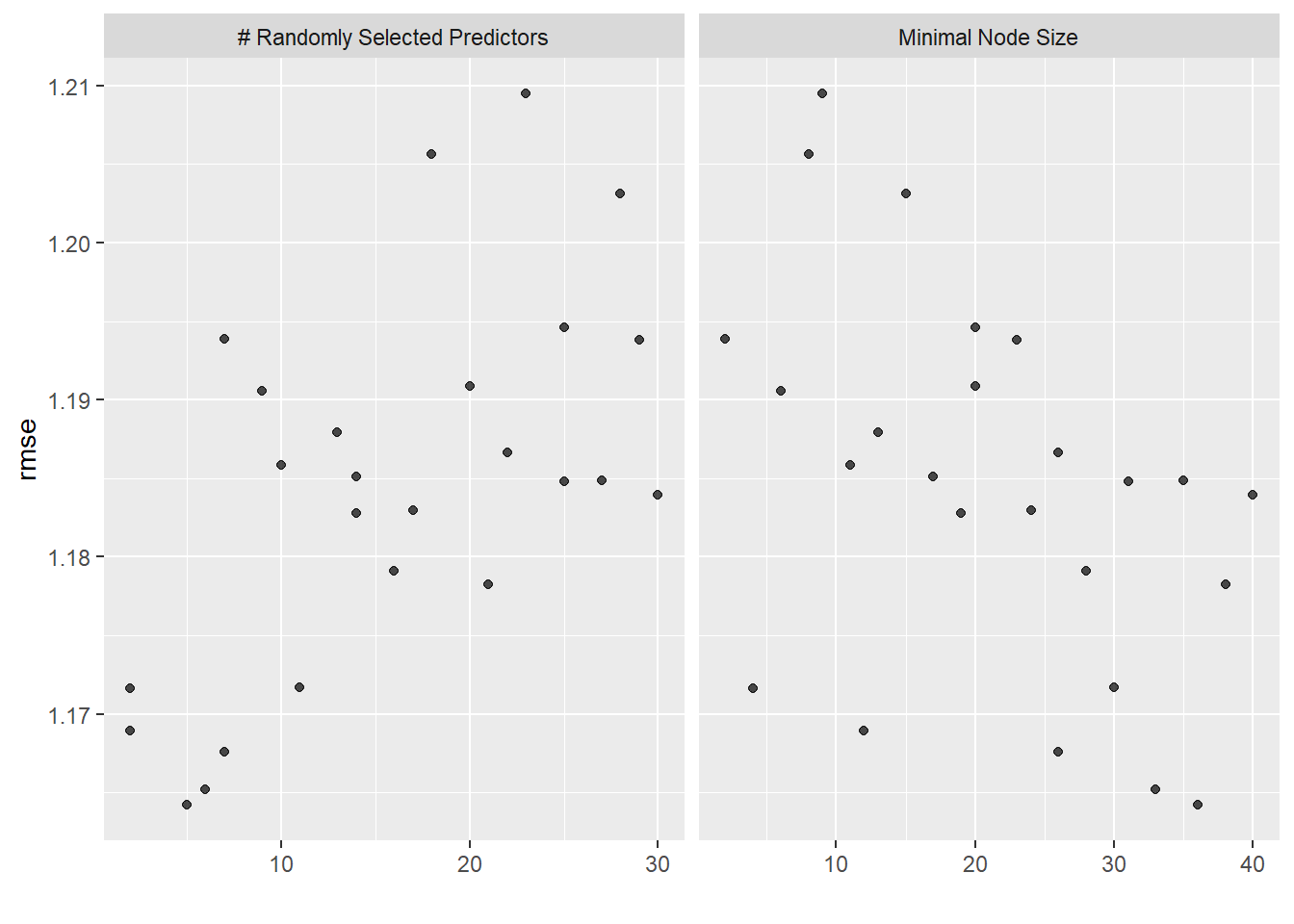
#getting best model
rf_res %>%
show_best(metric = "rmse")# A tibble: 5 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 5 36 rmse standard 1.16 25 0.0166 Preprocessor1_Model02
2 6 33 rmse standard 1.17 25 0.0165 Preprocessor1_Model23
3 7 26 rmse standard 1.17 25 0.0166 Preprocessor1_Model06
4 2 12 rmse standard 1.17 25 0.0167 Preprocessor1_Model25
5 2 4 rmse standard 1.17 25 0.0167 Preprocessor1_Model19best_rf <- rf_res %>%
select_best(metric = "rmse")
best_rf# A tibble: 1 × 3
mtry min_n .config
<int> <int> <chr>
1 5 36 Preprocessor1_Model02#finalizing workflow with best model
final_rf_wf <- rf_workflow %>%
finalize_workflow(best_rf)
final_rf_fit <- final_rf_wf %>% fit(data=flu_train1)
final_rf_fit══ Workflow [trained] ══════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: rand_forest()
── Preprocessor ────────────────────────────────────────────────────────────────
1 Recipe Step
• step_dummy()
── Model ───────────────────────────────────────────────────────────────────────
Ranger result
Call:
ranger::ranger(x = maybe_data_frame(x), y = y, mtry = min_cols(~5L, x), num.trees = ~1000, min.node.size = min_rows(~36L, x), num.threads = ~cores, verbose = FALSE, seed = sample.int(10^5, 1))
Type: Regression
Number of trees: 1000
Sample size: 508
Number of independent variables: 31
Mtry: 5
Target node size: 36
Variable importance mode: none
Splitrule: variance
OOB prediction error (MSE): 1.367179
R squared (OOB): 0.06700001 Model Evaluation
graphing actual vs predicted and residuals for each model
Tree
# get predicted and residual values in one dataset
## using augment() instead of predict() here so I can store everything in one df for easier graphing
tree_predict <- final_tree_fit %>%
augment(flu_train1) %>%
select(c(.pred, BodyTemp)) %>%
mutate(resid = BodyTemp - .pred)
tree_predict# A tibble: 508 × 3
.pred BodyTemp resid
<dbl> <dbl> <dbl>
1 99.2 97.8 -1.44
2 99.2 98.1 -1.14
3 98.7 98.1 -0.591
4 98.7 98.2 -0.491
5 98.7 97.8 -0.891
6 98.7 98.2 -0.491
7 98.7 98.1 -0.591
8 99.2 98 -1.24
9 99.2 97.7 -1.54
10 99.2 98.2 -1.04
# … with 498 more rows# Plot actual values vs predicted values
tree_pred_plot <- tree_predict %>%
ggplot(aes(x = BodyTemp, y = .pred)) +
geom_point() +
labs(title = "Predictions vs Actual",
x = "Body Temp Actual",
y = "Body Temp Prediction")
tree_pred_plot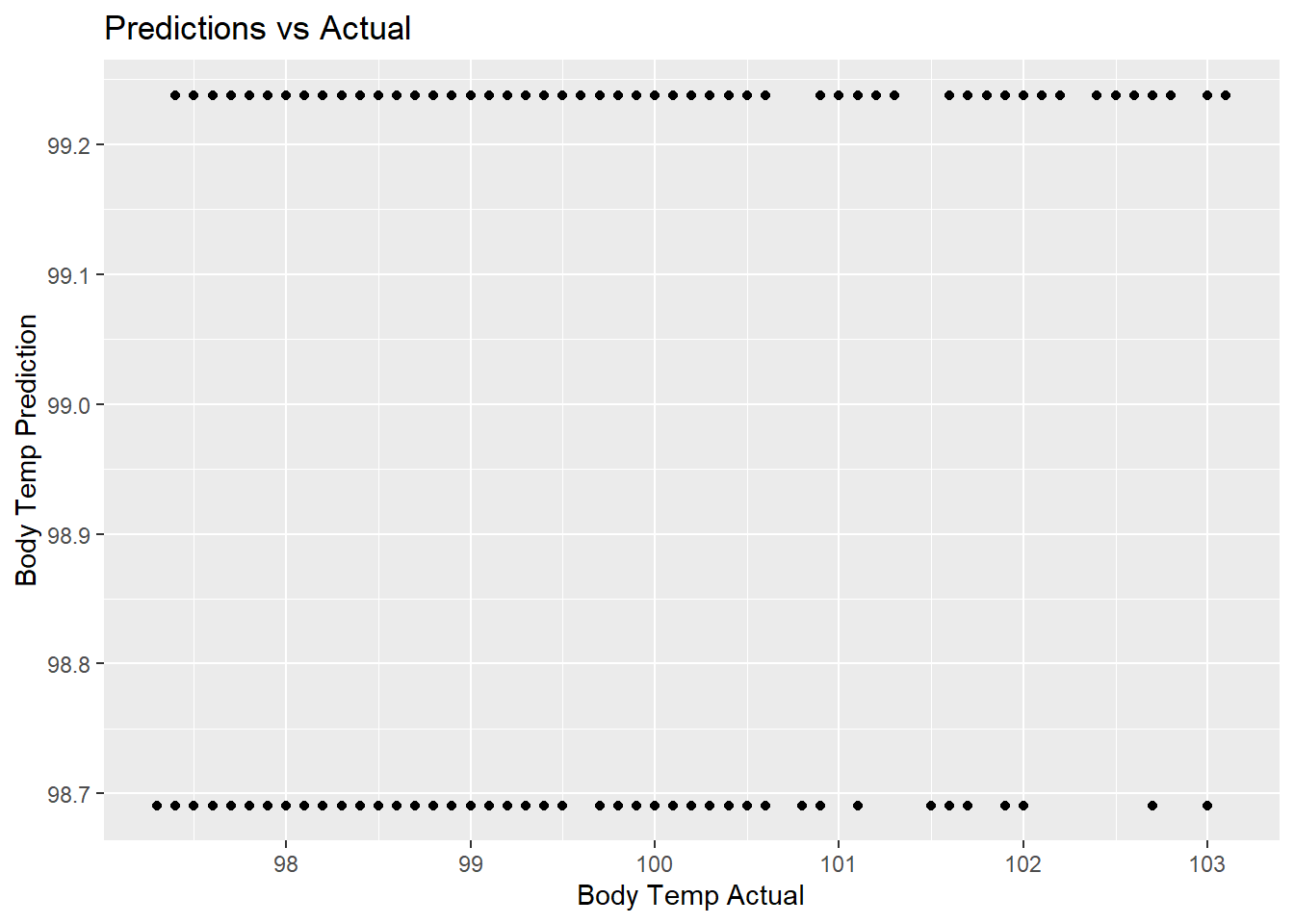
# Plot pred values vs residuals
tree_resid_plot <- tree_predict %>%
ggplot(aes(x = resid, y = .pred)) +
geom_point() +
labs(title = "Predictions vs Residual",
x = "Body Temp Residual",
y = "Body Temp Prediction")
tree_resid_plot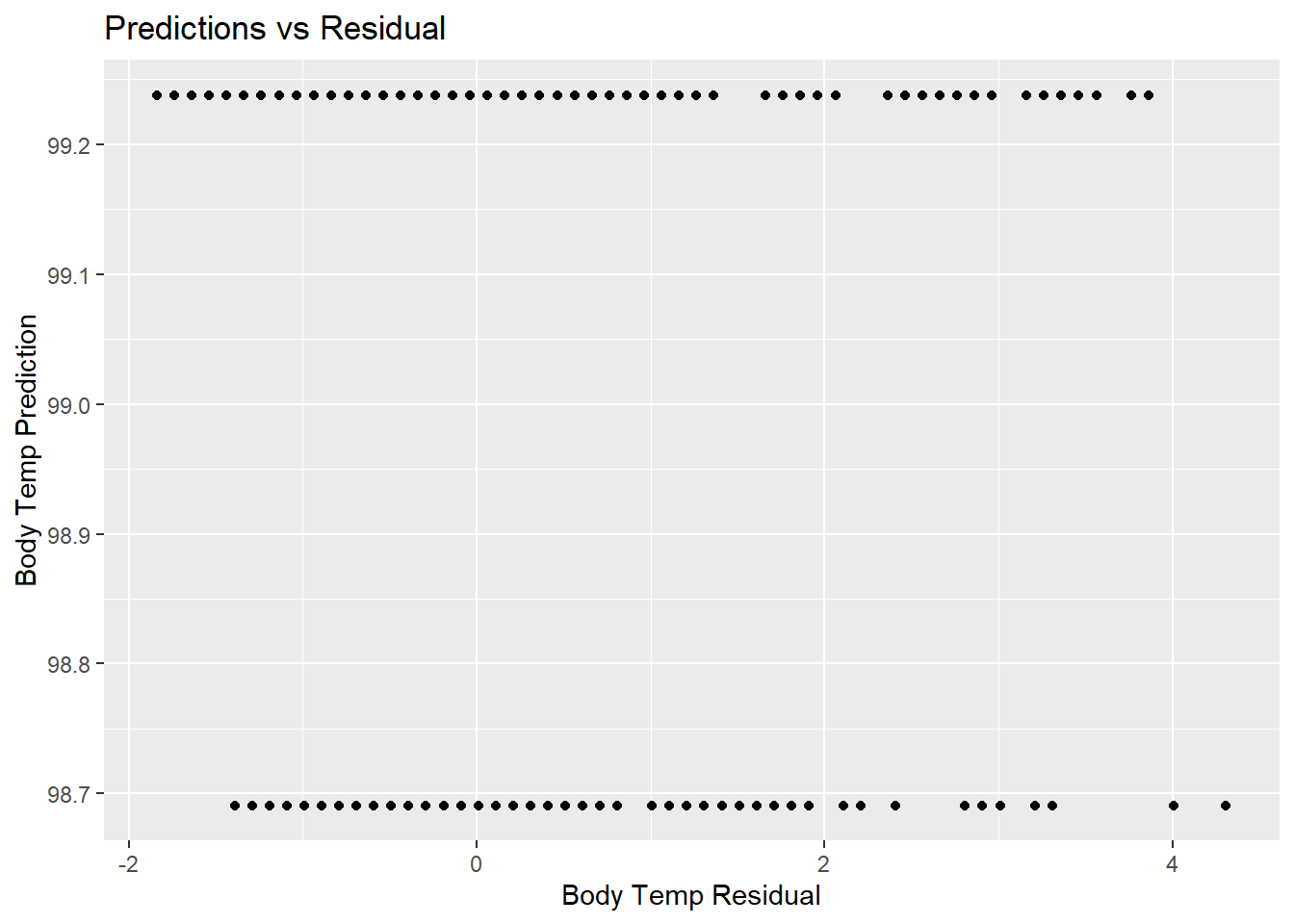
Lasso
# repeating the above process for lasso
# get predicted and residual values in one dataset
lr_predict <- final_lr_fit %>%
augment(flu_train1) %>%
select(c(.pred, BodyTemp)) %>%
mutate(resid = BodyTemp - .pred)
lr_predict# A tibble: 508 × 3
.pred BodyTemp resid
<dbl> <dbl> <dbl>
1 98.8 97.8 -0.950
2 98.8 98.1 -0.719
3 98.5 98.1 -0.360
4 98.8 98.2 -0.606
5 98.7 97.8 -0.907
6 98.7 98.2 -0.523
7 98.4 98.1 -0.257
8 99.3 98 -1.26
9 98.9 97.7 -1.24
10 99.0 98.2 -0.769
# … with 498 more rows# Plot actual values vs predicted values
lr_pred_plot <- lr_predict %>%
ggplot(aes(x = BodyTemp, y = .pred)) +
geom_point() +
labs(title = "Predictions vs Actual",
x = "Body Temp Actual",
y = "Body Temp Prediction")
lr_pred_plot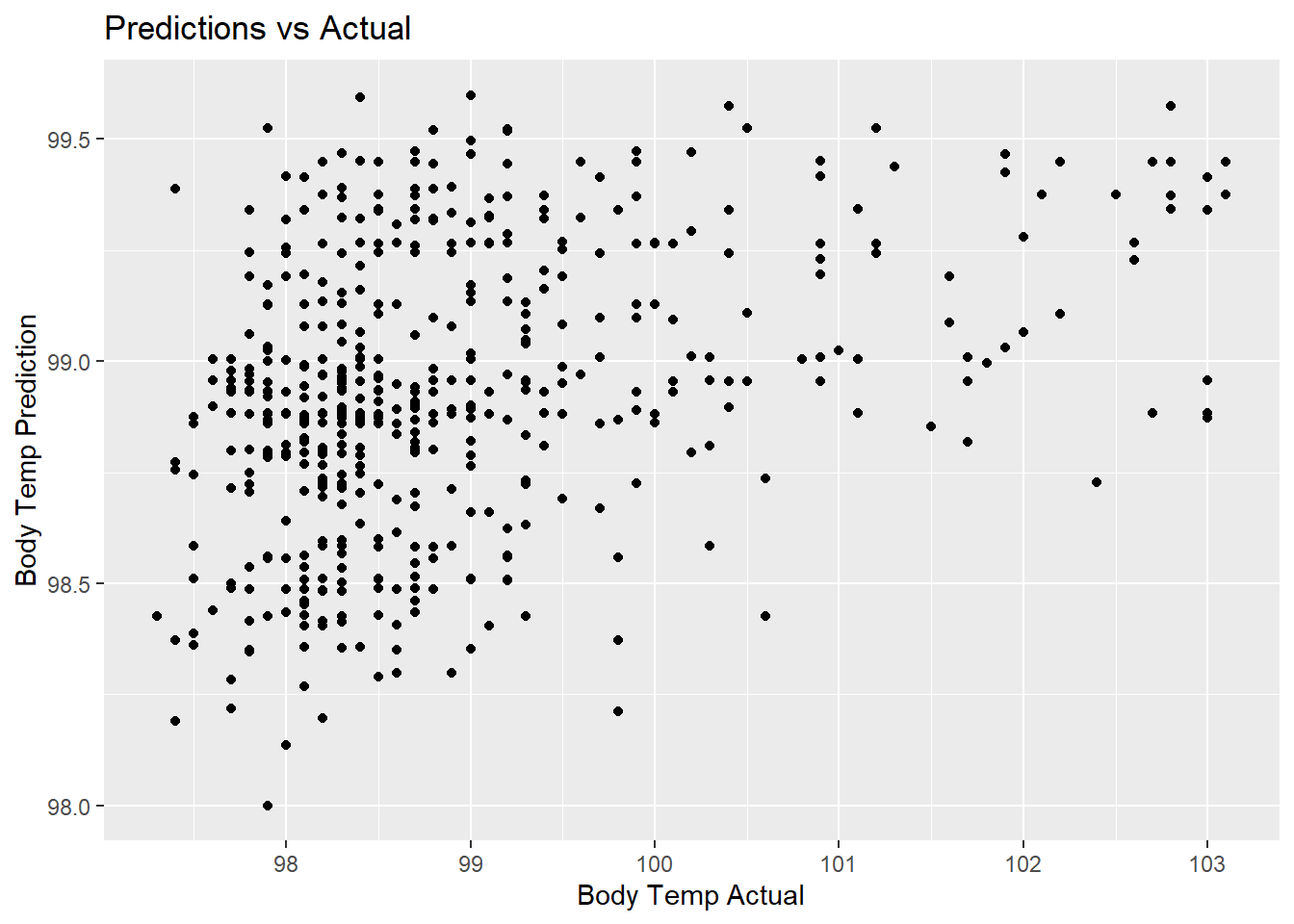
# Plot pred values vs residuals
lr_resid_plot <- lr_predict %>%
ggplot(aes(x = resid, y = .pred)) +
geom_point() +
labs(title = "Predictions vs Residual",
x = "Body Temp Residual",
y = "Body Temp Prediction")
lr_resid_plot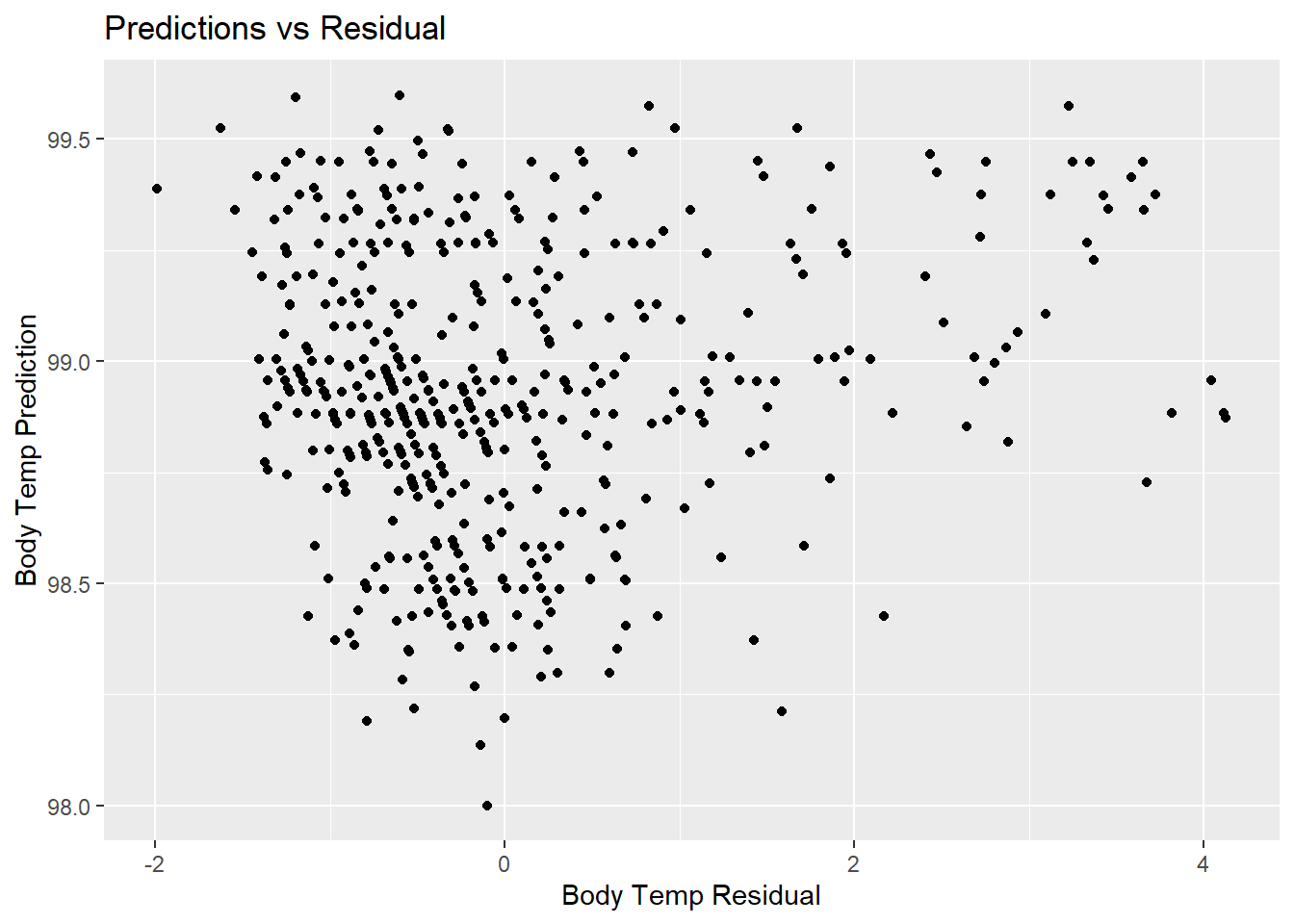
Random forest
# repeating again for random forest
# get predicted and residual values in one dataset
rf_predict <- final_rf_fit %>%
augment(flu_train1) %>%
select(c(.pred, BodyTemp)) %>%
mutate(resid = BodyTemp - .pred)
rf_predict# A tibble: 508 × 3
.pred BodyTemp resid
<dbl> <dbl> <dbl>
1 98.7 97.8 -0.929
2 98.6 98.1 -0.487
3 98.7 98.1 -0.595
4 98.7 98.2 -0.535
5 98.8 97.8 -0.998
6 98.6 98.2 -0.394
7 98.4 98.1 -0.256
8 99.1 98 -1.06
9 98.8 97.7 -1.06
10 98.9 98.2 -0.673
# … with 498 more rows# Plot actual values vs predicted values
rf_pred_plot <- rf_predict %>%
ggplot(aes(x = BodyTemp, y = .pred)) +
geom_point() +
labs(title = "Predictions vs Actual",
x = "Body Temp Actual",
y = "Body Temp Prediction")
rf_pred_plot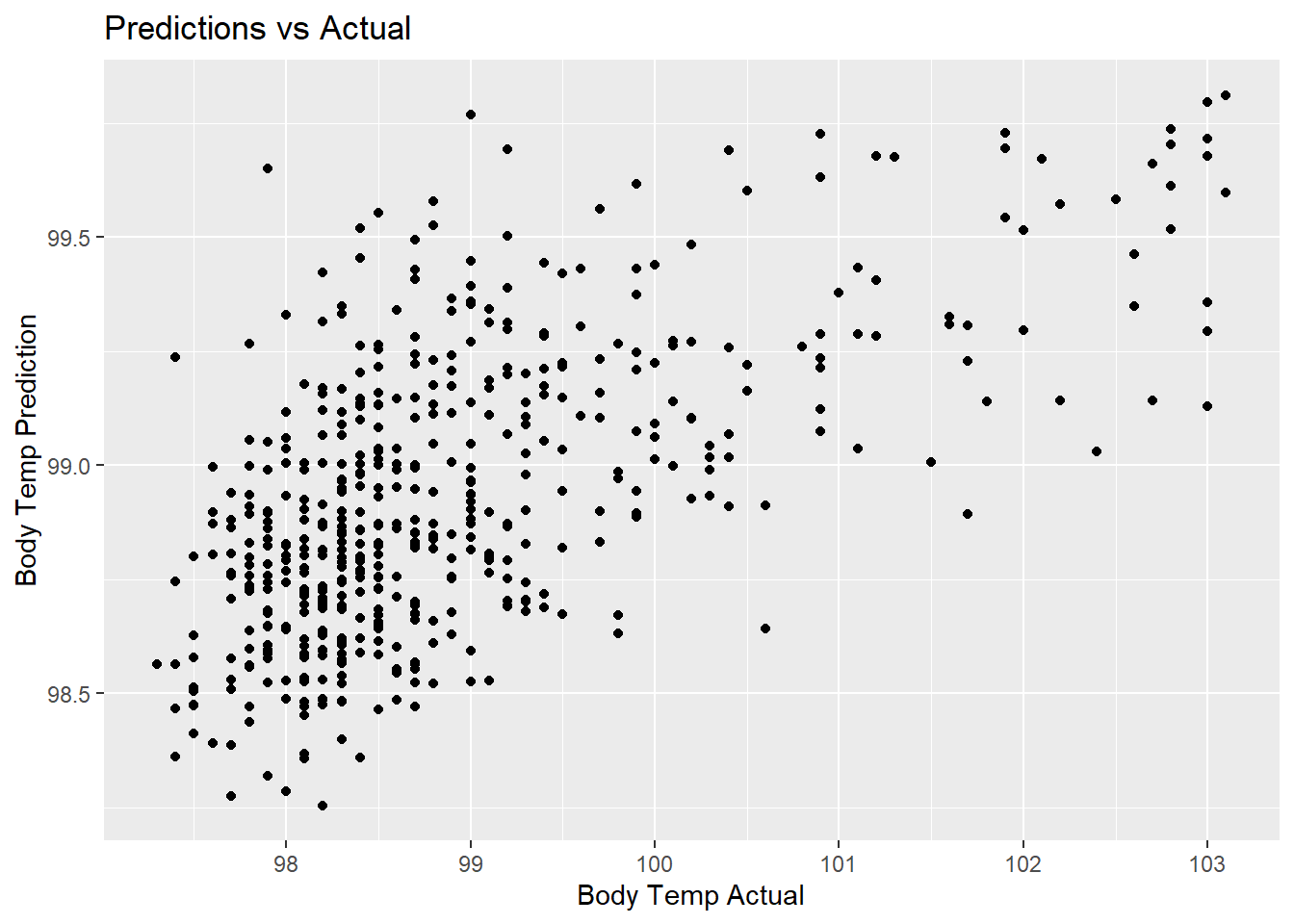
# Plot pred values vs residuals
rf_resid_plot <- rf_predict %>%
ggplot(aes(x = resid, y = .pred)) +
geom_point() +
labs(title = "Predictions vs Residual",
x = "Body Temp Residual",
y = "Body Temp Prediction")
rf_resid_plot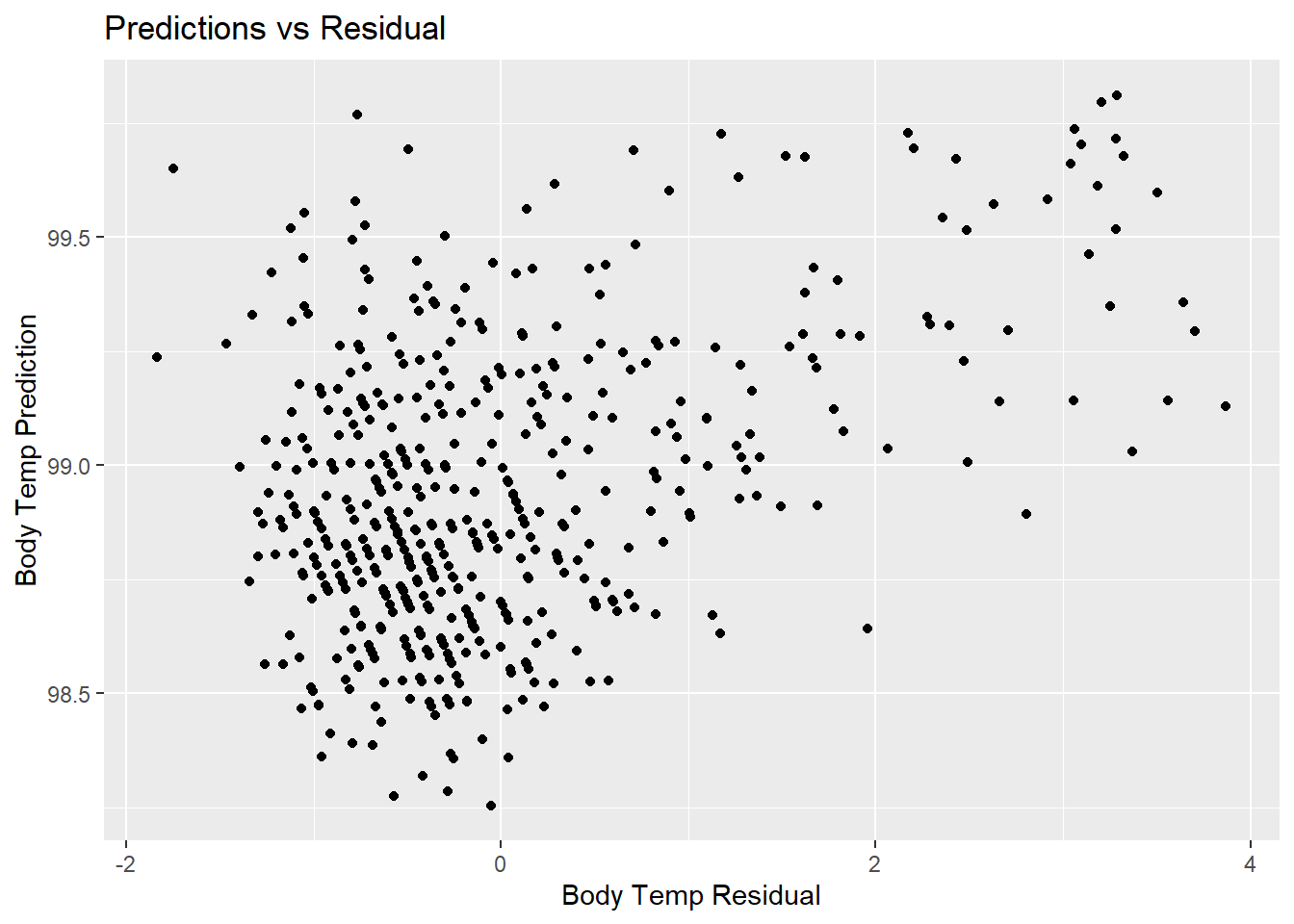
viewing performance
#comparing best models to null model to determine which model performed the best
tree_res %>%
show_best(metric = "rmse", n=1)# A tibble: 1 × 8
cost_complexity tree_depth .metric .estimator mean n std_err .config
<dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0000000001 1 rmse standard 1.19 25 0.0181 Preprocesso…lr_res %>%
show_best(metric = "rmse", n=1)# A tibble: 1 × 7
penalty .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0621 rmse standard 1.15 25 0.0169 Preprocessor1_Model28rf_res %>%
show_best(metric = "rmse", n=1)# A tibble: 1 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 5 36 rmse standard 1.16 25 0.0166 Preprocessor1_Model02null_train_fit %>%
collect_metrics(metric = "rmse")# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 1.21 25 0.0177 Preprocessor1_Model1
2 rsq standard NaN 0 NA Preprocessor1_Model1After evaluating all the models it appears that LASSO preformed the best as it has the lowest RMSE at 1.15 so I will be using it as my final model.
Final evaluation
#fitting lasso model to testing data with last_fit()
lr_last_fit <- final_lr_wf %>%
last_fit(split)
lr_last_fit %>% collect_metrics()# A tibble: 2 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 rmse standard 1.15 Preprocessor1_Model1
2 rsq standard 0.0291 Preprocessor1_Model1#includign null test metric for comparison
null_test_fit %>% collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 1.16 25 0.0285 Preprocessor1_Model1
2 rsq standard NaN 0 NA Preprocessor1_Model1I attempted the graphs but was throwing major errors trying to predict() or augment() based on a last_fit object. If I have time I will attempt this again but it will likely be later. Maybe someone else was successful and could provide some pointers?
The last fit returned an RMSE of 1.15341145 which is slightly higher than the best LASSO model above but better than the null model ran against the testing data.