Model Evaluation
Loading packages and cleaned data
# load packages library (tidyverse)
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.0 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.1 ✔ tibble 3.1.8
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.1
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
here() starts at C:/Users/Katie/Documents/2022-2023/MADA/katiewells-MADA-portfolio
── Attaching packages ────────────────────────────────────── tidymodels 1.0.0 ──
✔ broom 1.0.3 ✔ rsample 1.1.1
✔ dials 1.1.0 ✔ tune 1.0.1
✔ infer 1.0.4 ✔ workflows 1.1.3
✔ modeldata 1.1.0 ✔ workflowsets 1.0.0
✔ parsnip 1.0.4 ✔ yardstick 1.1.0
✔ recipes 1.0.5
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Learn how to get started at https://www.tidymodels.org/start/
# load cleaned data <- readRDS (here ("fluanalysis" , "data" , "flu2.rds" ))
Split data
set.seed (222 )# Put 3/4 of the data into the training set <- initial_split (flu2, prop = 3 / 4 )# Create data frames for the two sets: <- training (data_split)<- testing (data_split)
Create recipe
<- recipe (Nausea ~ ., data = train_data)
Fit model with recipe
#setting logistic regression engine <- logistic_reg () %>% set_engine ("glm" )
#creating workflow using model and recipe <- workflow () %>% add_model (lr_mod) %>% add_recipe (flu_rec)
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: logistic_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
0 Recipe Steps
── Model ───────────────────────────────────────────────────────────────────────
Logistic Regression Model Specification (classification)
Computational engine: glm
#make function to prepare the recipe and train the model <- %>% fit (data = train_data)
#pull fitting model object and make tibble of model coefficients %>% extract_fit_parsnip () %>% tidy ()
# A tibble: 32 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 2.56 9.39 0.273 0.785
2 SwollenLymphNodesYes -0.240 0.231 -1.04 0.300
3 ChestCongestionYes 0.168 0.252 0.668 0.504
4 ChillsSweatsYes 0.148 0.331 0.448 0.654
5 NasalCongestionYes 0.600 0.309 1.94 0.0521
6 SneezeYes 0.0974 0.247 0.395 0.693
7 FatigueYes 0.180 0.438 0.411 0.681
8 SubjectiveFeverYes 0.191 0.261 0.733 0.463
9 HeadacheYes 0.482 0.351 1.37 0.169
10 Weakness.L 0.668 0.406 1.65 0.0999
# … with 22 more rows
Using trained workflow to predict (test data)
#use trained model to predict with unseen data predict (flu_fit, test_data)
# A tibble: 183 × 1
.pred_class
<fct>
1 No
2 No
3 No
4 No
5 No
6 Yes
7 Yes
8 No
9 No
10 Yes
# … with 173 more rows
#save above together to use to get ROC <- augment (flu_fit, test_data)
#view the data %>% select (Nausea, .pred_No, .pred_Yes)
# A tibble: 183 × 3
Nausea .pred_No .pred_Yes
<fct> <dbl> <dbl>
1 No 0.962 0.0377
2 Yes 0.708 0.292
3 No 0.696 0.304
4 Yes 0.548 0.452
5 No 0.826 0.174
6 Yes 0.193 0.807
7 Yes 0.227 0.773
8 No 0.712 0.288
9 No 0.688 0.312
10 Yes 0.271 0.729
# … with 173 more rows
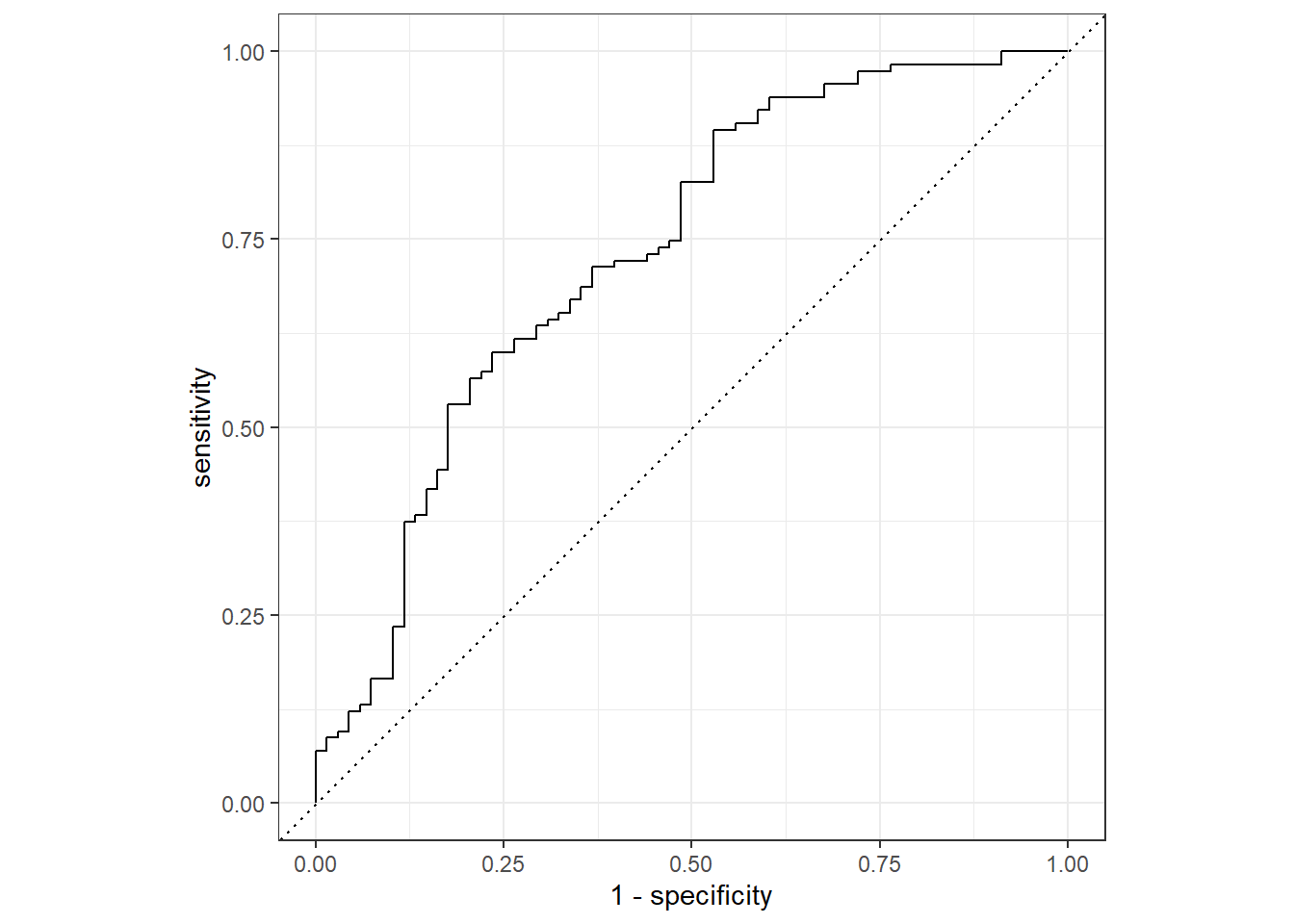
#create ROC curve with predicted class probabilities %>% roc_curve (truth = Nausea, .pred_No) %>% autoplot ()
Warning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
dplyr 1.1.0.
ℹ Please use `reframe()` instead.
ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
always returns an ungrouped data frame and adjust accordingly.
ℹ The deprecated feature was likely used in the yardstick package.
Please report the issue at <https://github.com/tidymodels/yardstick/issues>.
#ROC estimate %>% roc_auc (truth = Nausea, .pred_No)
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.731
Refitting with the main predicotr (RunnyNose) and using the trained model to predict with test and train data.
Alternative Model
<- recipe (Nausea ~ RunnyNose, data = train_data)
<- workflow () %>% add_model (lr_mod) %>% add_recipe (flu_rec2)
══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: logistic_reg()
── Preprocessor ────────────────────────────────────────────────────────────────
0 Recipe Steps
── Model ───────────────────────────────────────────────────────────────────────
Logistic Regression Model Specification (classification)
Computational engine: glm
<- %>% fit (data = train_data)
%>% extract_fit_parsnip () %>% tidy ()
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -0.790 0.172 -4.59 0.00000447
2 RunnyNoseYes 0.188 0.202 0.930 0.352
Using trained workflow to predict (test data)
predict (flu_fit2, test_data)
# A tibble: 183 × 1
.pred_class
<fct>
1 No
2 No
3 No
4 No
5 No
6 No
7 No
8 No
9 No
10 No
# … with 173 more rows
<- augment (flu_fit2, test_data)
%>% select (Nausea, .pred_No, .pred_Yes)
# A tibble: 183 × 3
Nausea .pred_No .pred_Yes
<fct> <dbl> <dbl>
1 No 0.688 0.312
2 Yes 0.646 0.354
3 No 0.646 0.354
4 Yes 0.646 0.354
5 No 0.688 0.312
6 Yes 0.688 0.312
7 Yes 0.646 0.354
8 No 0.688 0.312
9 No 0.688 0.312
10 Yes 0.688 0.312
# … with 173 more rows
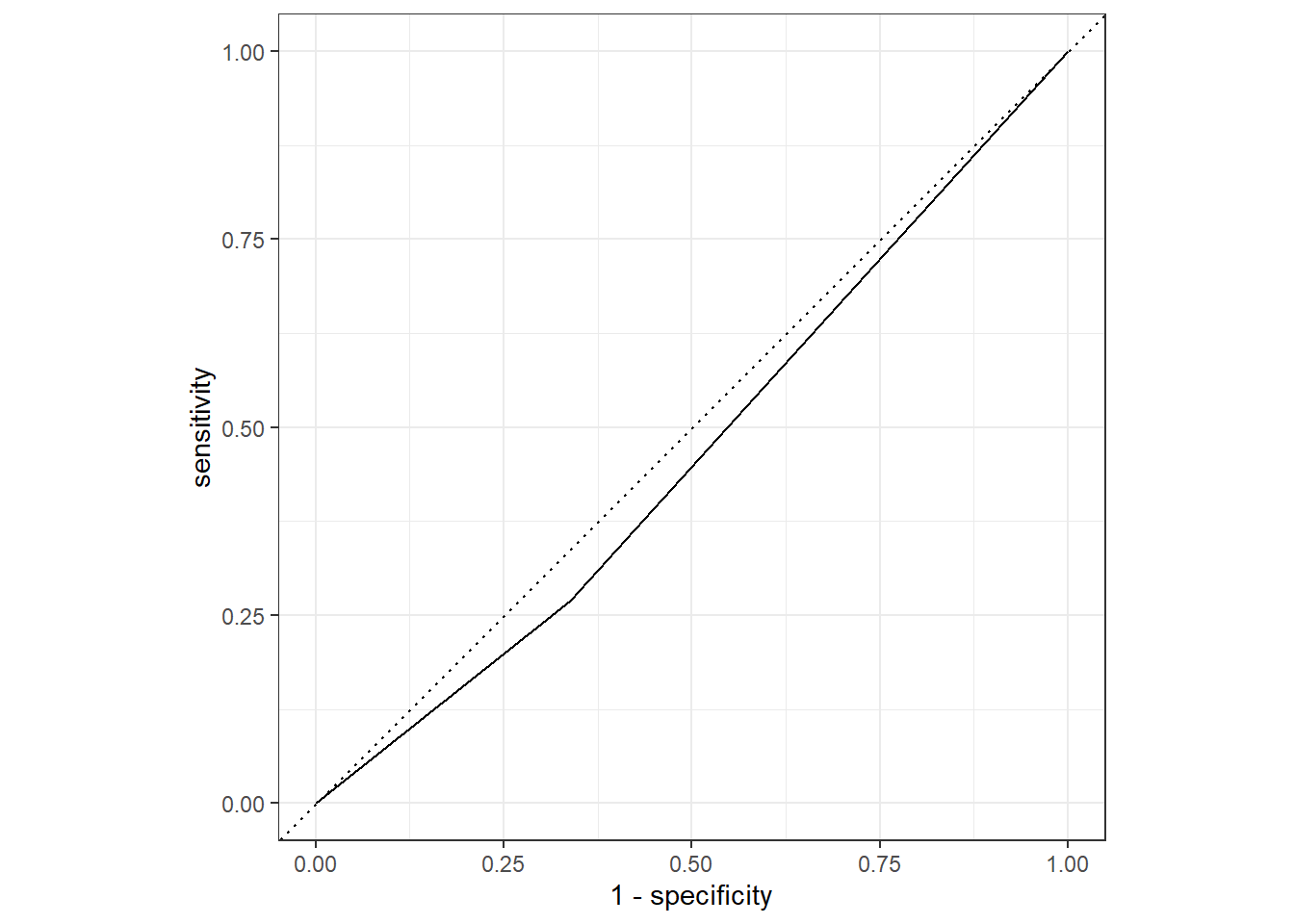
%>% roc_curve (truth = Nausea, .pred_No) %>% autoplot ()
%>% roc_auc (truth = Nausea, .pred_No)
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.466
The model using only runny nose as a predictor had a lower ROC than the model with all predictors.
This section added by Hayley Hemme
Create recipe
<- recipe (BodyTemp ~ ., data = train_data)
Fit linear model
#setting linear regression engine <- linear_reg () %>% set_engine ("lm" ) %>% set_mode ("regression" )
#creating workflow using model and recipe <- workflow () %>% add_model (ln_mod) %>% add_recipe (flu_ln_rec)
#make function to prepare the recipe and train the model <- %>% fit (data = train_data)
#pull fitting model object and make tibble of model coefficients %>% extract_fit_parsnip () %>% tidy ()
# A tibble: 32 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 98.0 0.280 350. 0
2 SwollenLymphNodesYes -0.190 0.108 -1.76 0.0789
3 ChestCongestionYes 0.146 0.115 1.27 0.203
4 ChillsSweatsYes 0.184 0.148 1.24 0.214
5 NasalCongestionYes -0.182 0.136 -1.34 0.180
6 SneezeYes -0.474 0.113 -4.18 0.0000338
7 FatigueYes 0.362 0.187 1.94 0.0529
8 SubjectiveFeverYes 0.564 0.118 4.79 0.00000223
9 HeadacheYes 0.0675 0.150 0.448 0.654
10 Weakness.L 0.224 0.182 1.23 0.218
# … with 22 more rows
Using trained workflow to predict (test data)
#use trained model to predict with unseen data predict (flu_ln_fit, test_data)
# A tibble: 183 × 1
.pred
<dbl>
1 99.4
2 99.0
3 99.8
4 98.7
5 99.0
6 99.6
7 99.2
8 98.8
9 99.5
10 98.8
# … with 173 more rows
#save above together <- augment (flu_ln_fit, test_data)
#view the data %>% select (BodyTemp, .pred)
# A tibble: 183 × 2
BodyTemp .pred
<dbl> <dbl>
1 98.3 99.4
2 98.8 99.0
3 102. 99.8
4 98.2 98.7
5 97.8 99.0
6 97.8 99.6
7 100 99.2
8 101. 98.8
9 98.8 99.5
10 100. 98.8
# … with 173 more rows
Measuring model fit - RMSE
%>% rmse (truth = BodyTemp, .pred)
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 1.15
Let’s also measure model fit using R2
%>% rsq (truth = BodyTemp, .pred)
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rsq standard 0.0446
Making a plot using predictions
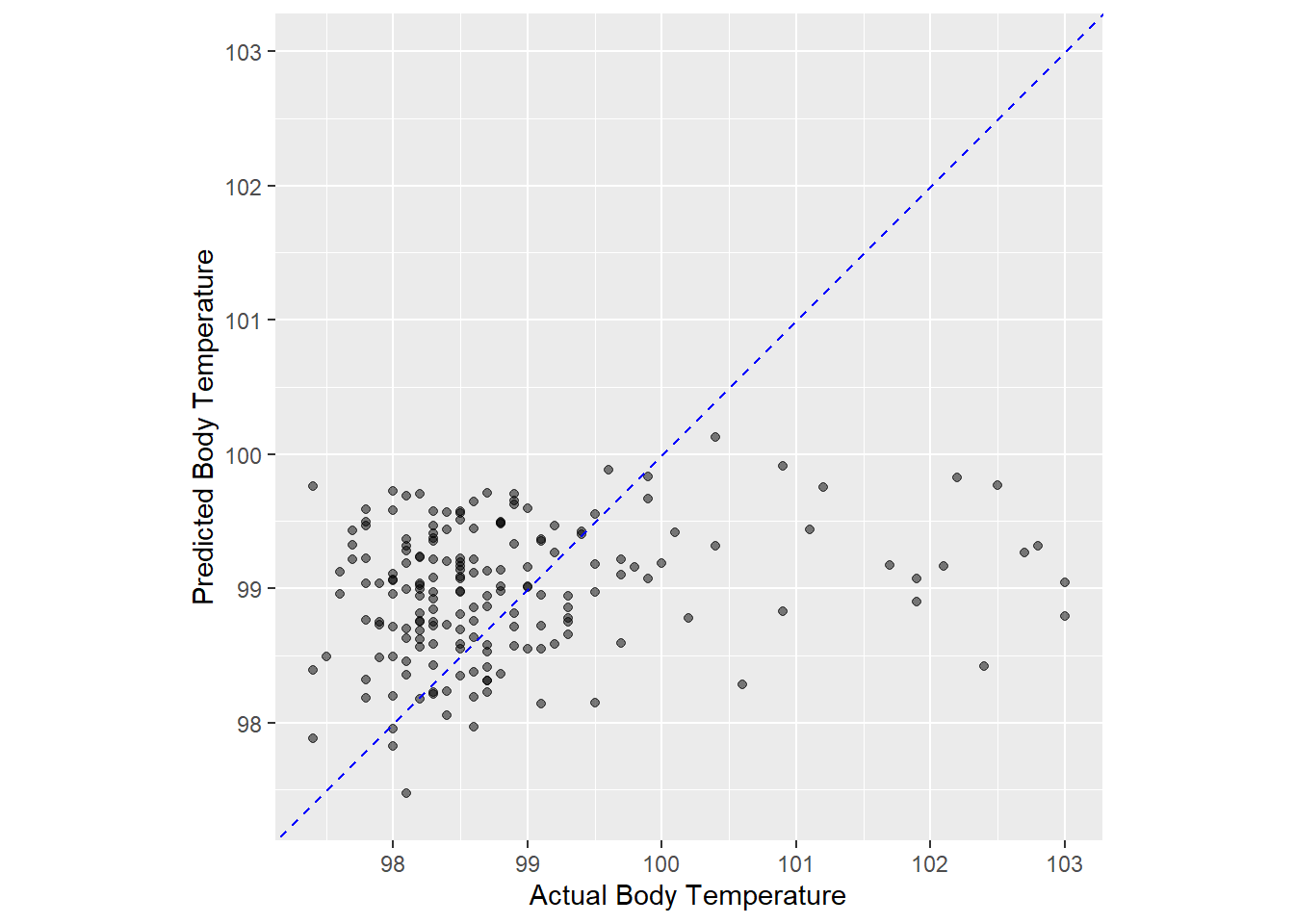
ggplot (flu_ln_aug, aes (x = BodyTemp, y = .pred)) + geom_point (alpha = 0.5 ) + geom_abline (color = 'blue' , linetype = 2 ) + coord_obs_pred () + labs (x = 'Actual Body Temperature' , y = 'Predicted Body Temperature' )
This trained model is not very well suited to predict body temperature using all the predictors in the model. Let move on and…
Refit with the main predictor (RunnyNose) and use the trained model to predict with test and train data. 11. Alternative Model Creating recipe
<- recipe (BodyTemp ~ RunnyNose, data = train_data)
Making workflow object
<- workflow () %>% add_model (ln_mod) %>% add_recipe (flu_ln_rec2)
Making fit object
<- %>% fit (data = train_data)
Extracting data of the model fit
%>% extract_fit_parsnip () %>% tidy ()
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 99.1 0.0964 1028. 0
2 RunnyNoseYes -0.261 0.114 -2.29 0.0225
Using trained workflow to predict (test data)
predict (flu_ln_fit2, test_data)
# A tibble: 183 × 1
.pred
<dbl>
1 99.1
2 98.9
3 98.9
4 98.9
5 99.1
6 99.1
7 98.9
8 99.1
9 99.1
10 99.1
# … with 173 more rows
<- augment (flu_ln_fit2, test_data)
%>% select (BodyTemp, .pred)
# A tibble: 183 × 2
BodyTemp .pred
<dbl> <dbl>
1 98.3 99.1
2 98.8 98.9
3 102. 98.9
4 98.2 98.9
5 97.8 99.1
6 97.8 99.1
7 100 98.9
8 101. 99.1
9 98.8 99.1
10 100. 99.1
# … with 173 more rows
Measuring model fit - RMSE
%>% rmse (truth = BodyTemp, .pred)
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 1.13
Let’s also measure model fit using R2
%>% rsq (truth = BodyTemp, .pred)
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rsq standard 0.0240
Make an R2 plot using predictions
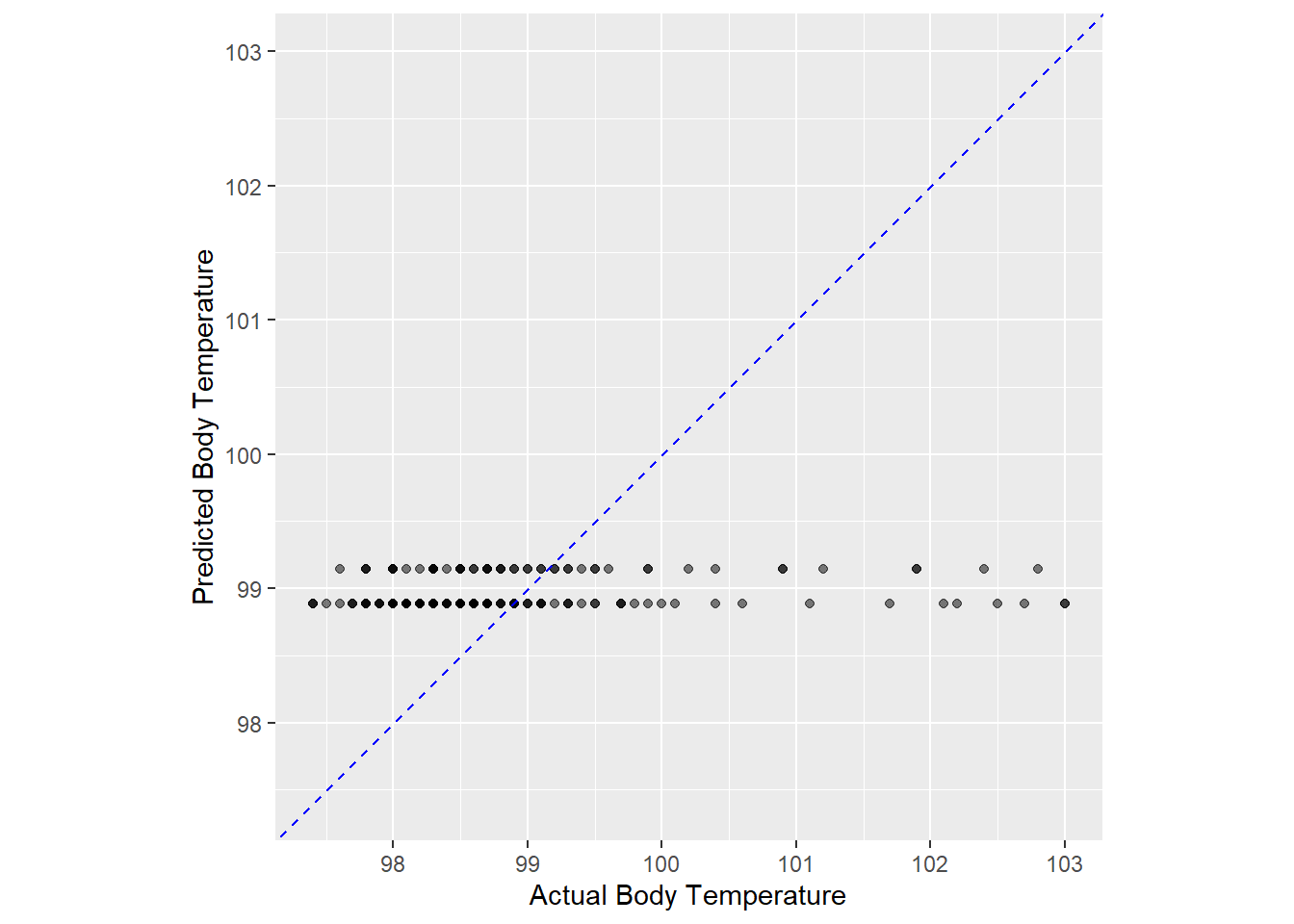
ggplot (flu_ln_aug3, aes (x = BodyTemp, y = .pred)) + geom_point (alpha = 0.5 ) + geom_abline (color = 'blue' , linetype = 2 ) + coord_obs_pred () + labs (x = 'Actual Body Temperature' , y = 'Predicted Body Temperature' )
The model preformed very poorly using runny nose as predictor of body temperature. I am also wondering why the model only had two different predictions, and makes me think that something may have went wrong with setting up the model?